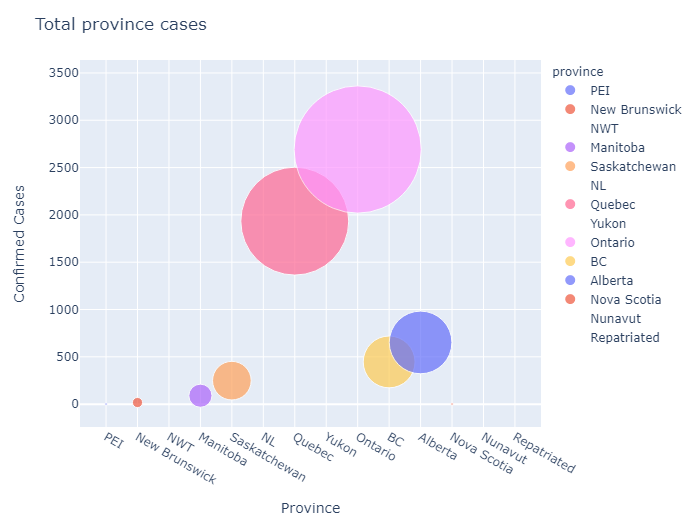
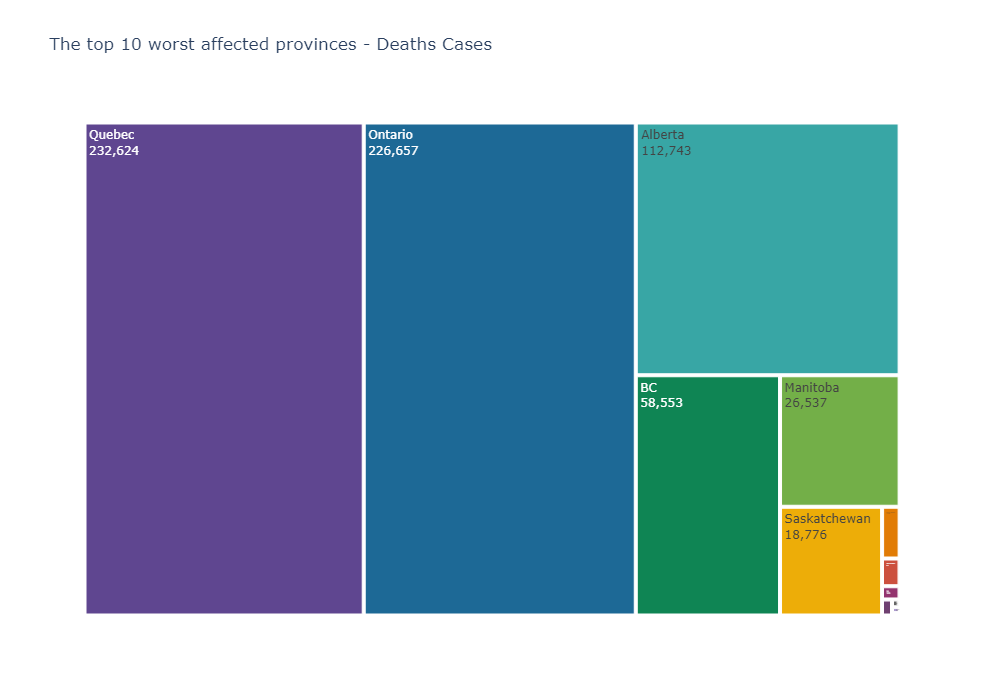
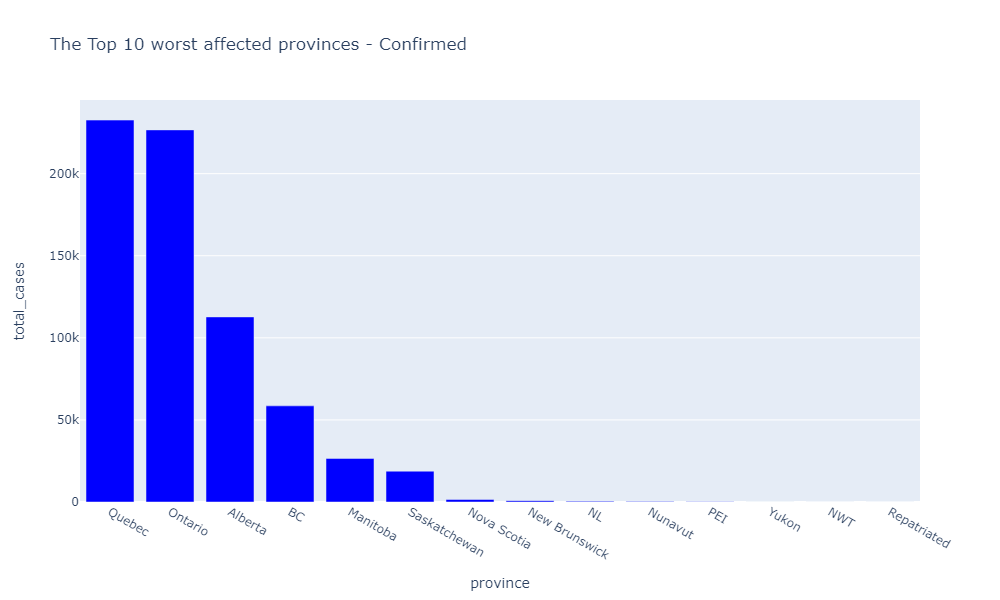
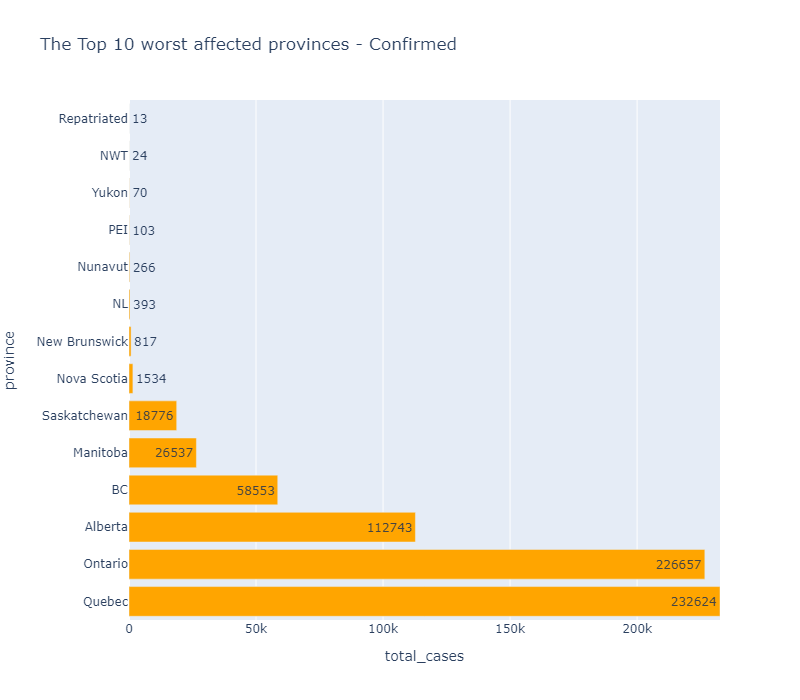
In [115]:
def covid_bubble_chart():
df = data_table_daily
fig = px.scatter(df, x="province", y="cases", size="cases", color="province",
hover_name="province", size_max=90)
fig.update_layout(
title="Total province cases",
xaxis_title="Province",
yaxis_title="Confirmed Cases",
width = 700
)
fig.show();
fig = go.FigureWidget( layout=go.Layout() )
interact(covid_bubble_chart)
ipywLayout = widgets.Layout(border='solid 2px green')
ipywLayout.display='none'
widgets.VBox([fig], layout=ipywLayout)